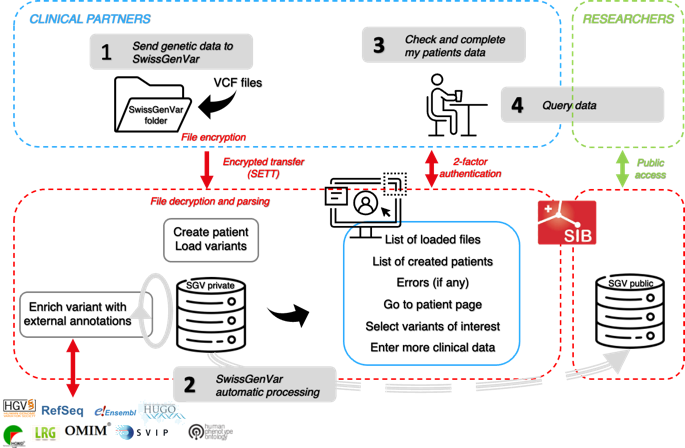
Application workflow #
Principle #
The project partners provide clinical grade data consisting in exome or genome sequencing data (in the form of VCF files) complemented with well characterized genetic and phenotypic information on the patients, entered by the partners directly on the SwissGenVar interface. In addition, for each variant, selected public information is automatically gathered on each variant, to support its interpretation.
Here is the summary of the data that will be collected by SwissClinGen:
| Data type | Entry format | Comment |
|---|---|---|
| Exome or genome data | VCF | SwissGenVar can already use SETT to transfer exomes from some clinical partners. |
| Variant information | Automatic retrieval by SwissGenVar | SwissGenVar already automatically retrieves selected public variants annotation, obtained from the VEP API. The number of data sources will be expanded during the project. |
| Curated clinical data | Manually entered in SwissGenVar | The clinical partners can add specific clinical phenotypic information for each patient, using the SwissGenVar interface. |
Data entry #
The genetic and clinical data are generated by the partner laboratories and can be provided either directly by the laboratory or the by hospital Clinical Data Warehouse (CDW), depending on each institution setup. In both cases, the data transfer takes place using the BioMedIT’s Secure Encryption and Transfer Tool “sett” . To date it has already been deployed and successfully used to transfer consented data from the Institute of Medical Genetics Zürich and the University Hospital of Geneva CDW, and the process in ongoing with Inselspital. We will continue deploying “sett” on all other partner sites.
In addition, for all variants in the VCF files, some public information listed in the Data section is already automatically gathered by the SwissGenVar platform using a local instance of the Variant Effect Predictor (VEP) deployed on the SENSA BioMedIT node. Other tools will be investigated for data not available in VEP.
Data query #
Users can retrieve patients or variants by querying the tool via its graphical interface, more details are provided in the following sections.
The Variant query page and the Patient query page are the following, respectively:
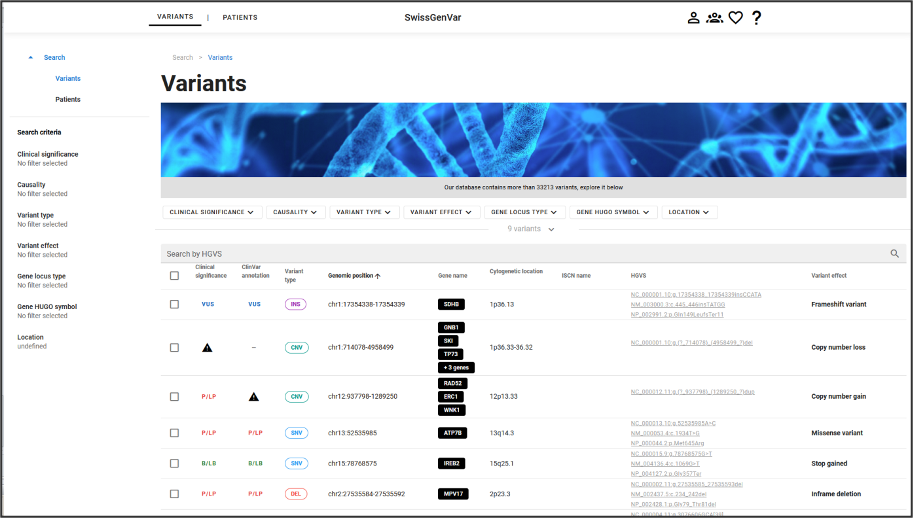
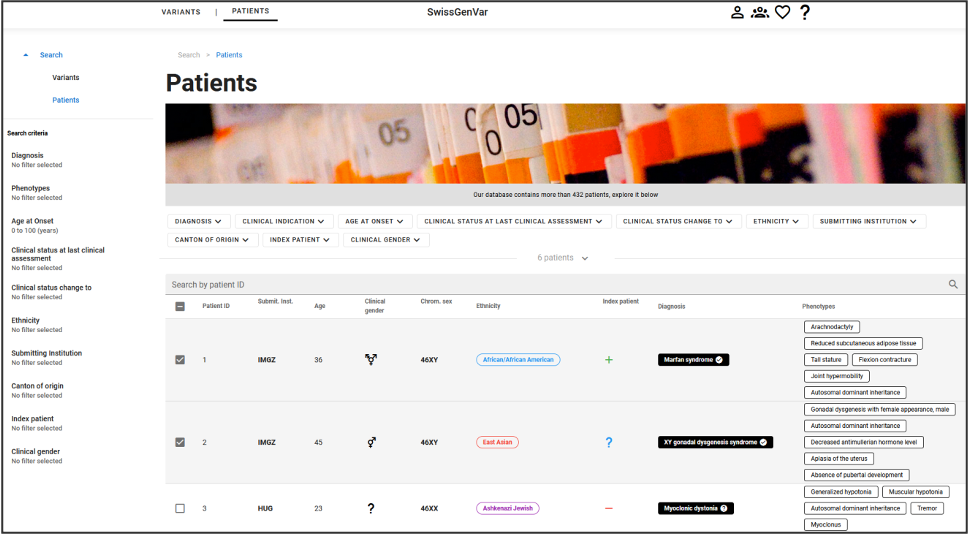
Those pages allow to search for specific information, that then can be looked at in more details.
Data display #
Once retrieved by a query, a single patient can be looked at, and its information can be completed with granular clinical information (see screenshot below). The list of varaints for this patient (obtained from the VCF) can be seen on the left of the page, while the right side comprises the phenotypic information.
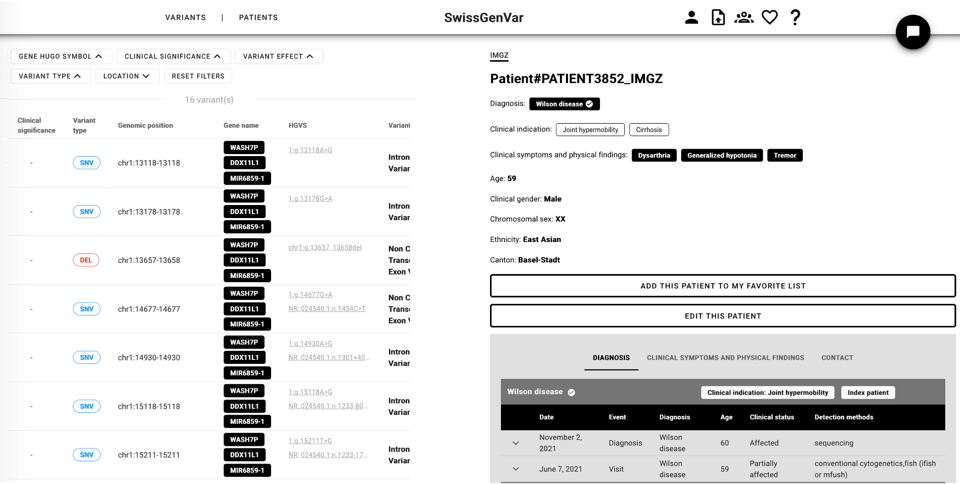
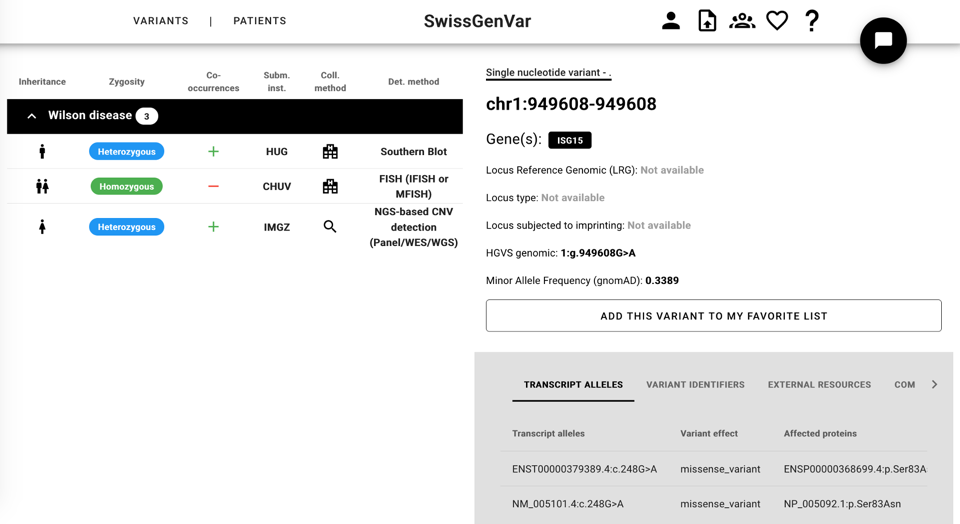
In the same manner, a single variant can be further looked at, and in this case the left side shows all the patients who present this variant in the database, while the right side shows the variant public information, automatically retrieved by SwissGenVar using VEP.
Global workflow schema #
The global workflow can be described as follows with 3 possible outputs.
graph TD A[START] --> B[SEARCH] A[START] --> C[ADD/EDIT] B -->|VARIANT| B1{SELECT FILTERS} B1 --> B11(LIST OF VARIANTS) B11 --> B12{SELECT A VARIANT} B12 --> B13(DETAILS OF VARIANT) B -->|PATIENT| B2{SELECT FILTERS} B2 --> B21(LIST OF PATIENTS) B21 --> B22{SELECT A PATIENT} B22 --> B23(DETAILS OF PATIENT) C --> C1{GENERAL INFO} C --> C2{GENETIC DATA} C --> C3{DIAGNOSIS/PHENOTYPES} C1 --> C11(FORM) C2 --> C21(UPLOAD VCF/MANUAL ENTRY) C3 --> C31(FORM) C11 --> C4{SHARE} C21 --> C4 C31 --> C4 C4 --> C5(PATIENT VISIBLE IN RESULTS) style A fill:#F44336 style B fill:#4CAF50 style C fill:#4CAF50 style B1 fill:#4CAF50 style B2 fill:#4CAF50 style B11 fill:#F44336 style B21 fill:#F44336 style B12 fill:#4CAF50 style B22 fill:#4CAF50 style B13 fill:#F44336 style B23 fill:#F44336 style C1 fill:#4CAF50 style C2 fill:#4CAF50 style C3 fill:#4CAF50 style C11 fill:#F44336 style C21 fill:#F44336 style C31 fill:#F44336 style C4 fill:#4CAF50 style C5 fill:#F44336
Query workflow #
graph TD A[START] --> B[SEARCH] B -->|VARIANT| B1{SELECT FILTERS} B1 --> B11(LIST OF VARIANTS) B11 --> B12{SELECT A VARIANT} B12 --> B13(DETAILS OF VARIANT) B -->|PATIENT| B2{SELECT FILTERS} B2 --> B21(LIST OF PATIENTS) B21 --> B22{SELECT A PATIENT} B22 --> B23(DETAILS OF PATIENT) style A fill:#F44336 style B fill:#4CAF50 style B1 fill:#4CAF50 style B2 fill:#4CAF50 style B11 fill:#F44336 style B21 fill:#F44336 style B12 fill:#4CAF50 style B22 fill:#4CAF50 style B13 fill:#F44336 style B23 fill:#F44336
Goal #
Create a query with one or multiple criteria to search the database and display the results corresponding to the query (variants or patients).
Description #
The query workflow is a search form which allows to build a query based on the user’s interests. Multiple criteria can be used to filter (see the filters section for more details).
Data flow #
Consented genetic data as well as manually curated data from genetic testing request forms or consultations can be entered into the SwissGenVar platform. Three types of data can be collected:
| Data type | Entry format | Comment |
|---|---|---|
| Exome or genome data | VCF | SwissGenVar can already use SETT to transfer exomes from some clinical partners. |
| Variant information | Automatic retrieval by SwissGenVar | SwissGenVar already automatically retrieves selected public variants annotation, obtained from the VEP API. The number of data sources will be expanded during the project. |
| Curated clinical data | Manually entered in SwissGenVar. | The clinical partners will add specific clinical phenotypic information for each patient, using the SwissGenVar interface. |
The data flows are represented on this scheme: